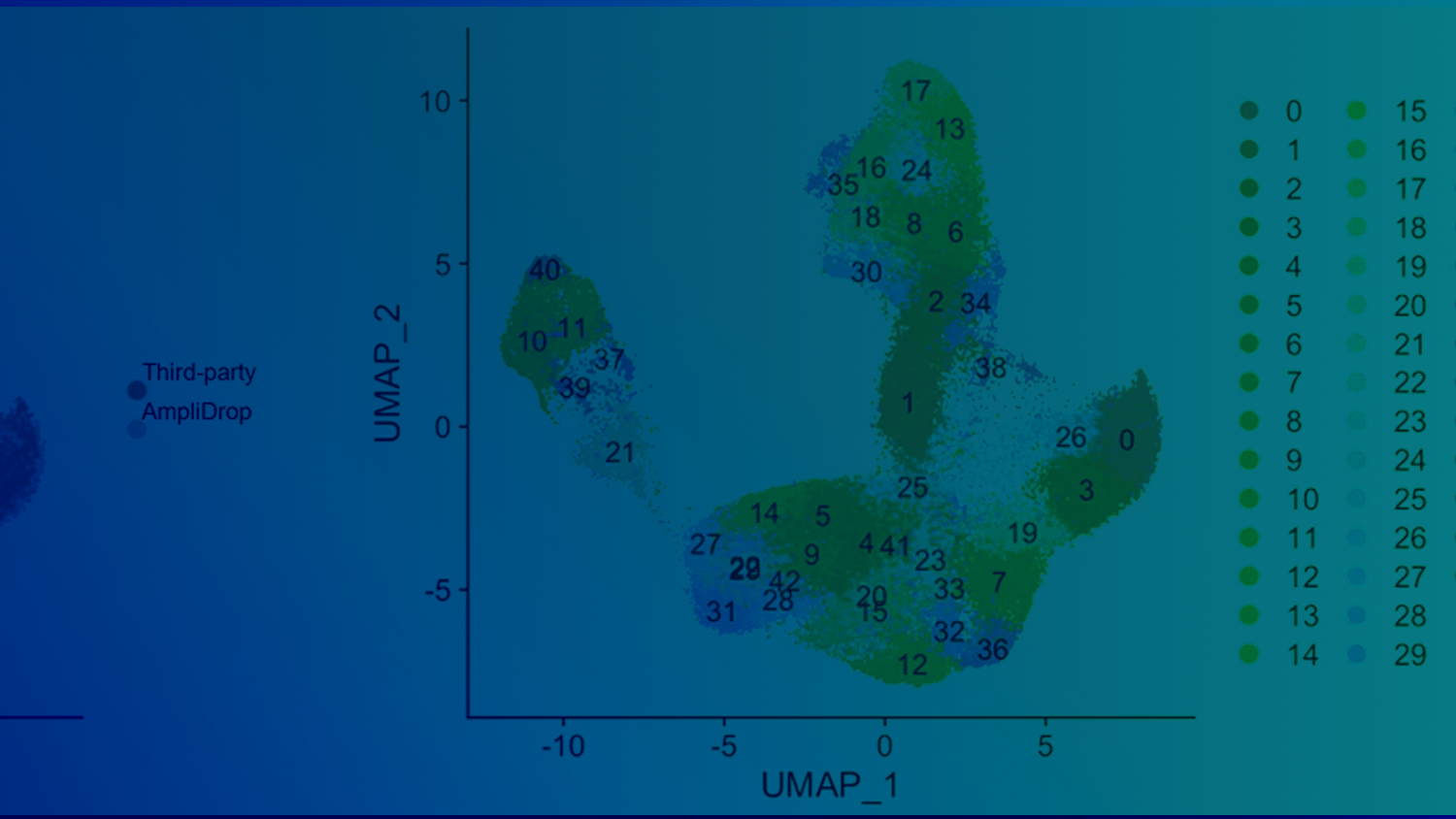
AmpliDrop™ Publications
Single-Molecule Barcoding Technology for Single-Cell Genomics (2024) (UST)
Single-Molecule Barcoding Technology for Single-Cell Genomics (2024) PDF
Ivan Garcia-Bassets, Guoya Mo, Yu Xia, Tsai-Chin Wu, Immanuel Mekuria, Veronika Mikhaylova, Madison Rzepka, Tetsuya Kawamura, Peter L. Chang, Amber Paasch, Long Pham, Surya Shiv Venugopal, Sandra Sanchez, Janaina S. de Souza, Likun Yao, Sifeng Gu, Zsolt Bodai, Alexis C. Komor, Alysson R. Muotri, Joy Wang, Yong Wang, Ming Lei, Angels Almenar-Queralt, and Zhoutao Chen
View in: BioRxiv
TREX1 is required for microglial cholesterol homeostasis and oligodendrocyte terminal differentiation in human neural assembloids (2024) (UST)
Gabriela Goldberg, Luisa Coelho, Guoya Mo, Laura A. Adang, Meenakshi Patne, Zhoutao Chen, Ivan Garcia-Bassets,
Pinar Mesci, Alysson R. Muotri
View: PubMed
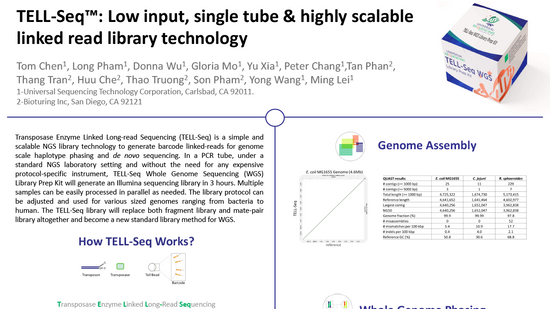
TELL-Seq™ Publications
High-quality genome assemblies for nine non-model North American insect species representing six orders (Insecta: Coleoptera, Diptera, Hemiptera, Hymenoptera, Lepidoptera, Neuroptera) (2024)
Kimberly K. O. Walden, Yanghui Cao, Christopher J. Fields, Alvaro G. Hernandez, Gloria A. Rendon, Gene E. Robinson, Rachel K. Skinner, Jeffrey A. Stein, Christopher H. Dietrich
View in: Molecular Ecology Resources
Low-cost and highly efficient generation of near-complete bacterial pathogen genomes byTELL-Seq (2024)
Low-cost and highly efficient generation of near-complete bacterial pathogen genomes by TELL-Seq PDF
Zhuoying Wu, Ting Zhang, Sheng Li, Lingzhu Shen, Zhangnv Yang,Yunfeng
Yang, Xuanheng Chen, Bolin Li, Shicong Zhou, Xintian Zhou, Beibei Wu, Jianmin
Jiang, Xiao Li
View in: bioRxiv
Targeted phasing of 2–200 kilobase DNA fragments with a short-read sequencer and a single-tube linked-read library method (2024) (UST)
Veronika Mikhaylova, Madison Rzepka, Tetsuya Kawamura, Yu Xia, Peter L. Chang, Shiguo Zhou, Amber Paasch, Long Pham, Naisarg Modi, Likun Yao,
Adrian Perez‑Agustin, Sara Pagans, T. Christian Boles, Ming Lei, Yong Wang, Ivan Garcia‑Bassets, Zhoutao Chen
View in: Nature
Characterizing the microbial metagenome of calcareous stromatolite formations in the San Felipe Creek in the Anza Borrego Desert (2024)
Rosalina Stancheva, Arun Sethuraman, Hossein Khadivar,
Jenna Archambeau, Ella Caughran, Ashley Chang, Brad Hunter, Christian Ihenyen, Marvin Onwukwe, Dariana Palacios, Chloe La Prairie, Nicole Read, Julianna Tsang, Brianna Vega, Cristina Velasquez, Xiaoyu Zhang, Elinne Becket, Betsy Read
View in: Genome Sequences
LRTK a platform agnostic toolkit for linked-read analysis of both human genome and metagenome (2024)
LRTK a platform agnostic toolkit for linked-read analysis of both human genome and metagenome (PDF)
Chao Yang, Zhenmiao Zhang, Yufen Huang, Xuefeng Xie,
Herui Liao, Jin Xiao, Werner Pieter Veldsman, Kejing Yin,
Xiaodong Fang, Lu Zhang
View: GigaScience
Anoxygenic phototroph of the Chloroflexota uses a type I reaction centre (2024)
Anoxygenic phototroph of the Chloroflexota uses a type I reaction centre (PDF)
J. M. Tsuji, N. A. Shaw, S. Nagashima, J. J. Venkiteswaran, S. L. Schiff, T. Watanabe,
M. Fukui, S. Hanada, M. Tank & J. D. Neufeld
View in: Nature
Exploring high-quality microbial genomes by assembling short-reads with long-range connectivity (2024)
Zhenmiao Zhang, Jin Xiao, Hongbo Wang, Chao Yang, Yufen Huang, Zhen Yue, Yang Chen, Lijuan Han, Kejing Yin, Aiping Lyu, Xiaodong Fang & Lu Zhang
View: Nature
Candida albicans selection for human commensalism results in substantial within-host diversity without decreasing fitness for invasive disease (2023)
Faith M. Anderson, Noelle D. Visser, Kevin R. Amses, Andrea Hodgins-Davis, Alexandra M. Weber, Katura M. Metzner, Michael J. McFadden, Ryan E. Mills, Matthew J. O’Meara, Timothy Y. James, Teresa R. O’Meara
View in: PLOS Biology
Understanding the impact of radical changes in diet and the gut microbiota on brain function and structure rationale and design of the EMBRACE study (2023)
Tair Ben-Porat, Angela Alberga, Marie-Claude Audet, Sylvie Belleville, Tamara R. Cohen, Pierre Y. Garneau, Kim L. Lavoie, Patrick Marion, Samira Mellah, Radu Pescarus, Elham Rahme, Sylvia Santosa, Anne-Sophie Studer, Dajana Vuckovic, Robbie Woods,
Reyhaneh Yousefi, Simon L. Bacon
Benchmarking genome assembly methods on metagenomic sequencing data (2023)
Benchmarking genome assembly methods on metagenomic sequencing data (2023)
Zhenmiao Zhang, Chao Yang, Werner Pieter Veldsman, Xiaodong Fang and Lu Zhang
View in: Briefings in Bioinformatics
Collapsible content
SpLitteR: Diploid genome assembly using TELL-Seq linked-reads and assembly graphs (2023)
SpLitteR: Diploid genome assembly using TELL-Seq linked-reads and assembly graphs (PDF)
IVAN TOLSTOGANOV, ZHOUTAO CHEN, PAVEL A. PEVZNER, ANTON KOROBEYNIKOV
View: BioRxiv (2023)
Genome Sequence of Lichtheimia ornata, an Emerging Opportunistic Mucorales Pathogen (2023)
Genome Sequence of Lichtheimia ornata, an Emerging Opportunistic Mucorales Pathogen (PDF)
TERRANCE SHEA, JASON T. MOHABIR, TANIA
KURBESSOIAN, BRITTANY BERDY, JAMES FONTAINE, ANDREAS GNIRKE, JONATHAN LIVNY,
JASON E. STAJICH, CHRISTINA A. CUOMO
View: Genome Sequences
Draft genome assemblies of the avian louse Brueelia nebulosa and its associates using long-read sequencing from an individual specimen (2023)
ANDREW D SWEET, DANIEL R BROWNE, ALVARO G HERNANDEZ, KEVIN P JOHNSON, STEPHEN L CAMERON AUTHOR NOTES
View: Oxford Academic (2023)
Genome of a novel Sediminibacterium discovered in association with two species of freshwater cyanobacteria from streams in Southern California (2022)
Whole-genome risk prediction of common diseases in human preimplantation embryos (2022)
Linked‑read sequencing for detecting short tandem repeat expansions (2022)
Linked‑read sequencing for detecting short tandem repeat expansions (PDF)
READMAN CHIU, INDHU-SHREE RAJAN-BABU, INANC BIROL & JAN M. FRIEDMAN
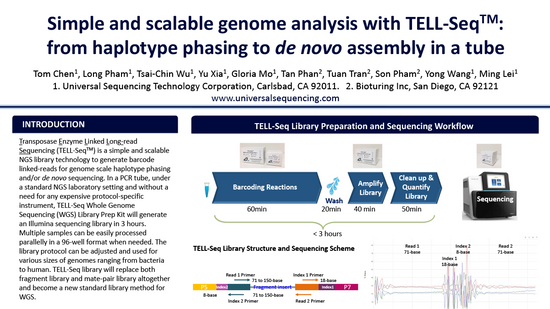
Ultra-low input single tube linked-read library method enables short-read second-generation sequencing systems to generate highly accurate and economical long-range sequencing information routinely (2000) (UST)
Zhoutao Chen, Long Pham, Tsai-Chin Wu, Guoya Mo, Yu Xia, Peter L. Chang,
Devin Porter, Tan Phan, Huu Che, Hao Tran, Vikas Bansal, Justin Shaffer, Pedro Belda-Ferre, Greg Humphrey, Rob Knight, Pavel Pevzner, Son Pham, Yong Wang, and Ming Lei
View in: Genome Research
cloudSPAdes: assembly of synthetic long reads using de Bruijn graphs (2019)
cloudSPAdes: assembly of synthetic long reads using de Bruijn graphs (PDF)
IVAN TOLSTOGANOV, ANTON BANKEVICH, ZHOUTAO CHEN, PAVEL A PEVZNER
View: Bioinformatics, Volume 35, Issue 14, July 2019, Pages i61–i70
Public Data Set
Ultra-low input single tube linked read library method enables short read NGS systems to generate highly accurate and economical long-range sequencing information...
Ultra-low input single tube linked read library method enables short read NGS systems to generate highly accurate and economical long-range sequencing information for de novo assembly and haplotype phasing